About the CoMSES Model Library more info
Our mission is to help computational modelers develop, document, and share their computational models in accordance with community standards and good open science and software engineering practices. Model authors can publish their model source code in the Computational Model Library with narrative documentation as well as metadata that supports open science and emerging norms that facilitate software citation, computational reproducibility / frictionless reuse, and interoperability. Model authors can also request private peer review of their computational models. Models that pass peer review receive a DOI once published.
All users of models published in the library must cite model authors when they use and benefit from their code.
Please check out our model publishing tutorial and feel free to contact us if you have any questions or concerns about publishing your model(s) in the Computational Model Library.
We also maintain a curated database of over 7500 publications of agent-based and individual based models with detailed metadata on availability of code and bibliometric information on the landscape of ABM/IBM publications that we welcome you to explore.
Displaying 5 of 15 results predator clear search
Soil microbe-predator model with enzymes
Randall Boone John C Moore Akihiro Koyama Kirstin Holfelder | Published Thursday, November 21, 2013We seek to improve understanding of roles enzyme play in soil food webs. We created an agent-based simulation of a simple food web that includes enzymatic activity. The model was used in a publication, Moore et al. (in press; Biochemistry).
Peer reviewed Ache hunting
Kim Hill Marco Janssen | Published Tuesday, August 13, 2013 | Last modified Friday, December 21, 2018Agent-based model of hunting behavior of Ache hunter-gatherers from Paraguay. We evaluate the effect of group size and cooperative hunting
Mast seeding model
Giangiacomo Bravo Lucia Tamburino | Published Saturday, September 08, 2012 | Last modified Saturday, April 27, 2013Purpose of the model is to perform a “virtual experiment” to test the predator satiation hypothesis, advanced in literature to explain the mast seeding phenomenon.
Landscape connectivity and predator–prey population dynamics
Jacopo A. Baggio | Published Thursday, November 10, 2011 | Last modified Saturday, April 27, 2013A simple model to assess the effect of connectivity on interacting species (i.e. predator-prey type)
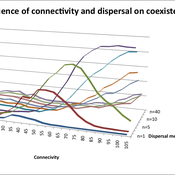
Varying effects of connectivity and dispersal on interacting species dynamics
Kehinde Salau | Published Monday, August 29, 2011 | Last modified Saturday, April 27, 2013An agent-based model of species interaction on fragmented landscape is developed to address the question, how do population levels of predators and prey react with respect to changes in the patch connectivity as well as changes in the sharpness of threshold dispersal?
Displaying 5 of 15 results predator clear search